Overview
The Wikipedia article describes proteins in general (quite a good description with links): http://en.wikipedia.org/wiki/Protein.
To introduce the description in this section, proteins are large molecules composed of a chain of small molecules (amino acids) that already exist. One (and sometimes several) gene(s) in the DNA determines the sequence of amino acids that compose the protein. Once created, the connected amino acid chain (polypeptide) is folded into its final shape, frequently combined with other proteins to form a final structure, and then transported to the location in the cell or body where it does its intended function.
Pretty well everything in a living body is composed of protein molecules. The human body contains approximately 50,000 different types of proteins: a human cell contains about 2 to 4 million proteins per cubic micron. To give an idea of the size of protein molecules, the largest known proteins are the titins, a component of the muscle sarcomere, with a molecular mass of almost 3,000 kDa (1 Da = the mass of 1 proton) and a total length of almost 27,000 amino acids. All proteins for an organism, as a group, are known as a proteome; there is an ongoing project to determine the type, number, function, and other characteristics of all proteins (see http://www.peptideatlas.org/publications/PA_JPR_2013.pdf for an introduction).
The body (cell) is the location that creates new proteins. The process of creating proteins involves a vast number of existing proteins, specifically engineered for that task. Most descriptions are simplified to identify only the key proteins required.
Initiating the creation of a protein
When the body senses that it requires a type of protein, it sends a signal protein to the cell(s) that are to generate that protein. The signal protein enters the cell and starts a process that "recruits" the other types and quantities of proteins required to locate the gene on the DNA. Those existing helper proteins are called transcription factors and there are about 1500 different types of transcription factor proteins in a human cell.
Finding the gene
How does the initial signal protein end up "finding" the exact gene in the DNA that it needs in order to create the proper template for the intended protein?
Over the 10s of million years, organisms have evolved a very complicated many-layered, but effective, way of finding a specific target gene from among the 20-25,000 possible targets and 3 million or so other elements of the DNA in the chromatin. The transcription factors are able to be tuned to detect particular anomalies in the DNA chain that identify more specific contents along the DNA. The transcription factor proteins twirl along exposed DNA strands until they encounter identifiers that prompt the factors to jump elsewhere to continue the search. Judging from all that I've been reading, transcription factors probably move along a DNA strand at about 30 base pairs a second. DNA strands tightly twirled around histone protein complexes loosen if protein factors indicate their desire to explore those areas. Histones in areas of interest can be "rolled" along as to unwind DNA lengths of interest. With many protein factors searching for the same gene(s), the target genes can be found quickly.
Although the formula "transcription factors find gene, polymerase creates mRNA protein template for that gene" is standard, there are many ways in which the cell can do this. Life is fascinating, but not simple.
Creating the mRNA protein template
Once the location(s) are found on the DNA strand, a polymerase complex quickly assembles and starts the transcription process. This protein complex rushes along the DNA strand and unzips the two connected strands in the DNA. The complex attracts nucleotides from the chromatin and attaches the corresponding nucleotide to each exposed half of the now-split base pair, thus creating a mirror of one of the two strands. Once the mirror strand (called messanger RNA (mRNA)) is complete, it and the polymerase complex detach.
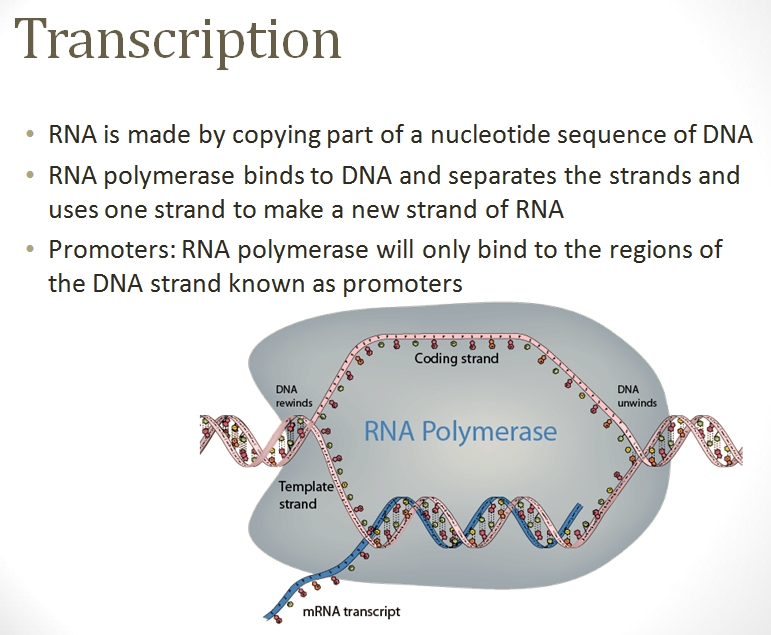
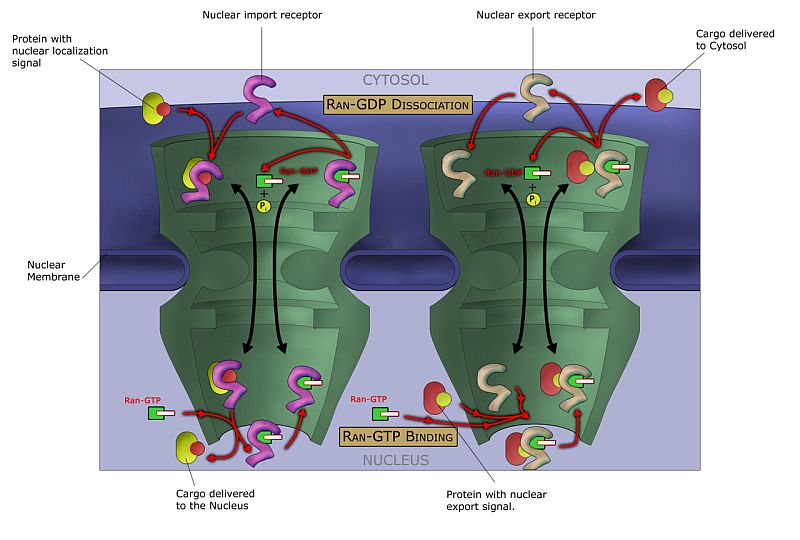
The mRNA that are to exit the nucleus attract protein complexes called exportins, which guide the mRNA molecule through the nucleus pores and into the cell cytoplasm.
The mRNA meets the ribosome
?
Translating the mRNA into the protein chain
Three items are necessary to translate the mRNA code (the sequence of 3 nucleotides (codons) along the mRNA molecule) into protein: the mRNA molecule, the ribosome protein complex, and 1000s of "charged" transfer RNA (tRNA) molecules. "Charged" in this case means that the tRNA molecule has had an amino acid added to its pointy end. That amino acid is intended to detach and become part of (be transfered to) any new protein being created by a ribosome.
The flat end (anticodon) of the tRNA molecule contains 3 out of 4 possible nucleotides, meaning that there are 24 possible tRNA molecules. The empty tRNA molecules are transcripted in the nucleus and exit into the cytoplasm. There, the enzyme aminoacyl tRNA synthetase "charges" each empty tRNA molecule with the amino acid that corresponds to the 3-part anticodon. Because only 20 amino acid types are available, some tRNAs carry duplicate amino acids.
The cytoplasm is rich in tRNA molecules. The ribosome protein complex (composed of some 13 proteins) attracts thousands of tRNA molecules into it, one at a time. The ribosome is mechanically structured to contain 3 cavities. The tRNA molecules enter from one side, are pushed into the center. If the tRNA anticodon corresonds to the 3 complimentary nucleotides currently in the ribosome (the codon), the ribosome attracts an ATP energy molecule, which, by splitting into an ADP and releasing a phosphate group and an electron, supplies the ribosome with the energy to move the amino acid from the end of the tRNA and attach the acid molecule to the end of the growing chain of amino acids (polypeptide) that are forming the new protein. Then the now-empty tRNA is pushed into the exit cavity and then out of the ribosome, and the ribosome moves along the mRNA to the next set of 3 nucleotides.
Once the amino acids become part of the polypeptide chain, the amino acids are called amino acid residues.
Folding the protein chain into its final shape
As the protein chain is being created, the electromagnetic and chemical properties of the amino acid residues attract and repel sections of the same chain so that they stick together (fold) in various ways. The folding process can start while the protein chain is birthing out of the ribosome and can continue all the way to when the protein reaches its functional destination. Frequently, existing proteins control the degree to which, when, and how a protein chain folds. The final shape of the protein determines its overall electromagnetic, chemical, and mechanical properties.
RNA transcription and folding
In addition to regular protein transcription and translation, the cell can create types of RNA molecules that fold and act as proteins in various cell processes (for example, such as tRNA). For processes requiring much energy quickly, the cell can use the RNA-creation process to create functional molecules at the rate and quantity required.
Creating complex proteins
Transporting the protein to its destination
Examples, uses, and specific proteins
Protein death, and recycling
Once a protein has completed its job in a cell, the protein is typically disposed of to ensure that it does not continue to function when no longer needed, and to recycle its component amino acids. The proteasome is a multiprotein structure that cells employ to destroy and recycle unwanted proteins. Several molecules specifically tag proteins destined for destruction. Without the tag molecules specifically designed for such destined-for-death proteins, useful proteins might be degraded as well.